Evolution of Plant-Pathogen Associations

In Australia, studies of the ecological and evolutionary dynamics of host-pathogen systems have centred on empirical studies of metapopulations of Linum marginale subject to attack by the rust pathogen Melampsora lini. Extensive investigations have also been undertaken of two rust pathogen-plant interactions in island archipelagoes off the Swedish coast (Filipendula ulmaria-Triphragmium ulmariae; Valeriana salina-Uromyces valerianae). We have recently initiated long-term studies on a fourth plant host-pathogen interaction, the coastal vagrant Cakile maritima and it's pathogen, Alternaria brassicicola. This system provides a useful contrast to the Linum, Filipendula and Valeriana systems in that resistance in Cakile is likely to be quantitative rather than gene-for-gene.
The Linum marginale-Melampsora lini System
Detailed information on variation in host resistance and pathogen infectivity has been difficult to obtain for many host-pathogen systems regardless of whether they are plant or animal based. The genetically best-documented interaction is the classic 'gene-for-gene' association occurring between the rust pathogen Melampsora lini and its agricultural host Linum usitatissimum first elucidated in the 1940s. The naturally occurring interaction between the endemic Australian species Linum marginale and M. lini is related to, but evolutionarily distinct from that between the pathogen and its agricultural host. Over the past 15 years, studies have focused on understanding the epidemiology, resistance and infectivity structure of this naturally occurring host-pathogen system. Much of this work has been centred within a single well-defined metapopulation, with a focus on coevolutionary dynamics across multiple spatial scales.
Within single populations, extensive long-term host demographic and disease monitoring has demonstrated the highly stochastic nature of the amplitude of disease incidence and prevalence from year-to-year, with high mortality associated with severe epidemic conditions. Populations of L. marginale are frequently composed of many different host resistance phenotypes distributed unevenly through the deme; sympatric pathogen populations are also typically diverse although not infrequently they may be dominated by just a few pathotypes. Year-to-year fluctuations in the incidence and frequency of different pathotypes provides strong circumstantial evidence for the effects of population bottlenecks and subsequent genetic drift. Analysis of multi-year data on disease prevalence has provided the first evidence that observed levels of asynchrony in disease dynamics may indeed be related to resistance structure in nature.

At the metapopulation level, recent surveys of multiple host and pathogen populations have revealed significant spatial structure in the distribution of resistance such that host populations in close proximity were more closely related than more distantly placed ones. In contrast, reflecting the pathogen's greater mobility, relatively little structure was detected at the same spatial scale in pathogen populations. Characterisation of patterns of local adaptation in the Linum-Melampsora system illustrates that apparent levels of local adaptation of pathogens can vary unpredictably among populations depending on overall levels of host resistance, suggesting an important role for local history. Our results also show how the coevolutionary interaction between hosts and pathogens can be further influenced by local structure such that resistant hosts select for generally virulent pathogens, while susceptible hosts select for more avirulent pathogens, strongly suggesting the possibility of an evolutionary trade-off between infectivity and virulence in this system.
At larger geographic scales, comparisons between adjacent metapopulations have shown that differences in host mating system affect the way in which variation for resistance is partitioned within and among individuals and populations. These differences are likely to have a significant impact on the spatial patterning of resistance phenotypes within populations. The consequences of such differences in structure for the epidemiology of the disease have yet to be assessed.
Overall, our work demonstrates considerable variability in the resistance and infectivity structure of host and pathogen populations. However, the tight frequency-dependent cycling predicted by early gene-for-gene models is not evident at the individual population level, and raises several important questions which are the focus of our current work: (i) Does a lack of correspondence between resistance and infectivity genes at the local scale imply similar patterns at regional scales? (ii) How does the persistence of gene-for-gene interactions (or indeed genetic polymorphisms in general) depend on spatial structure? (iii) Can spatial structure promote the persistence of gene-for-gene interactions regionally, even if such interactions break down locally due to stochastic extinction and colonisation events?
Scientific Staff: Burdon, Thrall
TOP
The Cakile maritima-Alternaria brassicicola System


The Cakile maritima-Alternaria brassicicola system differs from the Linum-Melampsora interaction in several major ways that make comparative studies of particular value (e.g. quantitative vs. gene-for-gene resistance). Because Cakile only grows on beach foreshores, it is easy to delineate populations, making this system ideal for spatial studies. The study area includes 60 populations in three subregions along the New South Wales south coast.
A primary goal is to examine how spatial structure relates to disease dynamics, in the context of physical factors that relate to long-term plant and pathogen persistence. Disease dynamics in the subregions are similar, but disease severity varies significantly among these areas, indicating the presence of large-scale environmental factors influencing timing and development of epidemics.
Disease Prevalence Across the Metapopulation
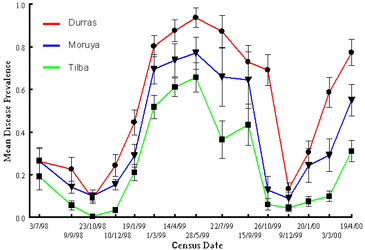
Variation in disease dynamics among individual beaches depends on physical factors that influence exposure to the sea, and thus host and pathogen persistence across growing seasons (e.g. population extinction is more likely on beaches with greater wind exposure and sea access).
Because among-population movement of seeds is by ocean current, beaches with greater sea access are also more likely to be recolonised. The potential for vertical transmission of Alternaria through infected seeds means that sea access is also likely to play a role in pathogen spread among host populations. Overall, significant levels of colonisation and extinction, together with the implication that connectedness among populations is likely to be distance dependent suggests the Cakile-Alternaria interaction exhibits dynamics typical of a metapopulation.
A major goal for studies of the Cakile-Alternaria system is to develop an understanding of within- and among-population variation in host resistance and to ask whether this is linked to overall differences in disease dynamics in the different subregions. This will involve conducting genetic studies to determine if resistance is indeed quantitatively-based in this system, and assessing the degree to which pathogens are locally adapted to their host populations. A second major goal is to evaluate the extent to which there is spatial structure in pathogen populations, as well as overall levels of variation in pathogen populations. Initial results using AFLPs indicate strong isolation by distance effects within a single subregion; an ongoing project centres around spatial structure among subregions. While there are good reasons to suspect that the evolutionary dynamics of hosts and pathogens would differ in quantitative and gene-for-gene systems, to date no study has examined resistance and infectivity structure in a way that allows meaningful comparisons. Because we will have long-term data on colonisation and extinction, we will also be able to explicitly ask how variation in host and pathogen population structure relates to dynamics within the Cakile-Alternaria metapopulation.
Scientific Staff: Burdon, Thrall
TOP
Theoretical Studies of Plant-Pathogen Interactions
This project focuses on modelling interactions between plants and pathogens
in spatially realistic situations. Dispersal ability clearly impacts on
the spatial structure of host-pathogen interactions, but may also influence
the evolution of resistance and infectivity. We initially developed a
genetically simplified, but otherwise biologically realistic metapopulation
model in which host and pathogen dispersal distances could be varied to
examine effects on pathogen persistence and dynamics. The spatial scale
of pathogen dispersal was an important factor in determining the probability
of disease persistence, as well as spatial and temporal patterns of incidence
and prevalence.
Disease Dynamics and Persistence
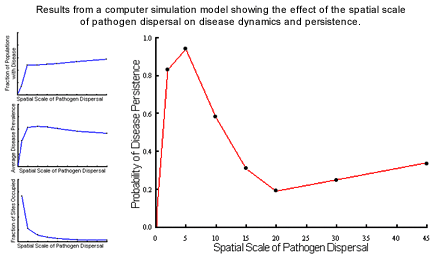
Disease persistence was highest at relatively local scales of dispersal where
colonisation and extinction processes play significant roles in the interaction
(most typical of natural systems); at higher scales disease persistence
was less likely due to major reductions in the fraction of sites occupied
by the host. The nature of the dynamics also varied with spatial structure,
with local pathogen dispersal leading to endemic dynamics while more global
dispersal resulted in epidemic cycles typical of aerially-dispersed rusts.
We are extending this work to investigate gene-for-gene interactions with a more realistic model that assumes 5 resistance and infectivity loci (32 possible genotypes) and again includes within and among-population demographic processes, but also allows mutation at individual loci, and among-population gene-flow. This model is being used to specifically ask how altering spatial structure through dispersal influences the coevolutionary dynamics of hosts and pathogens, in particular the overall levels of diversity that evolve, and the potential for maintaining genetic polymorphisms. Our simulations demonstrate clear impacts of dispersal distance on the total number of host and pathogen genotypes that evolve and are maintained in the metapopulation, as well as the number of resistance or infectivity genes carried by individual hosts or pathogens.
TOP
Evolutionary Dynamics of Gene-For-Gene Systems
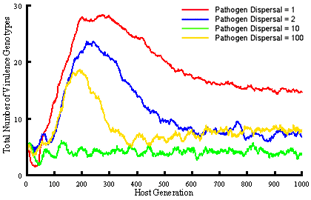
Major conclusions to date are: (i) pathogen 'super-races' do not always evolve and become dominant (indeed, it is not necessary to assume costs of resistance or infectivity to maintain genetic polymorphisms), (ii) rates of coevolution depend on the spatial extent of dispersal, with greater mixing resulting in a faster approach to equilibrium, (iii) at dispersal scales where colonisation and extinction play a role, evolutionary outcomes differ from situations where dispersal is either very local or global. It is at these scales where the diversity of host and pathogen genotypes is greatest.
A major issue in host-pathogen interactions is to identify conditions that favour gene-for-gene resistance/infectivity systems vs. those that favour more quantitatively-based systems. Examples of such variation exist within single host-pathogen systems, suggesting that shifts in the ecology of the interaction may favour different genetic structures, however there is little data, and no theoretical studies which have addressed this issue. Future research will centre on development of metapopulation models based on quantitative variation in resistance and infectivity (parallel to the gene-for-gene models). Similar to the studies discussed above, this model will then be used to investigate disease dynamics and host-pathogen coevolution in relation to spatial structure.
Scientific Staff: Burdon, Thrall