Ecology & Genetics of Remnant Vegetation
Habitat fragmentation is a major threat to many of Australia's indigenous
plant species. Understanding its biological effects
is crucial to the effective management of remnant plant populations for
conservation. In addition, maintenance of remnant vegetation within agricultural
landscapes is critical for its contribution to production through the
provision of ecosystem services. The aims of this research project are
to quantify how genetic and demographic processes interact to determine
the viability of native plant populations occupying fragmented habitats.
Of particular interest is the assessment of the roles of inbreeding and
genetic erosion in limiting performance of a range of ecologically differing
target species. This research will provide tests of current conservation
theory regarding genetic and demographic effects of fragmentation, as
well as generating specific management guidelines for the species being
studied.
The research employs a broad range of techniques including the use of
molecular genetic markers, demographic monitoring, growth experiments
and simulation modelling to examine various aspects of the genetics and
ecology of remnant populations. Much of the work described below is reviewed
in: Young A.G. & Clarke G.M. (eds)
Genetics, Demography and the Viability of Fragmented Populations
(2000) Cambridge University Press and Young A.G. et al. (eds) Forest
Conservation Genetics: Principles and Practice, CSIRO Publishing.
Ecological Genetics of Endangered Plants

The temperate grassland communities of the southern tablelands in New South Wales and the ACT have been largely destroyed by land clearing, erosion, changing water tables and invasive plants and animals since European settlement some 200 years ago. While previously occupying some 0.5 million ha, the 10 000 ha that currently remains is now distributed across the landscape as a mosaic of patches separated by improved pasture. These remnants vary in size, degree of isolation and, perhaps most importantly, level of degradation. Despite this, they remain one of the most floristically diverse plant communities in Australia and contain high numbers of both endangered and threatened plant species. This research project investigates how genetic and demographic processes interact to determine the viability of rare and endangered plant species within this grassland community. A primary aim is to use this information to improve species conservation management plans.
Several species are currently being studied. These have been chosen to represent contrasting life history types within the grassland plant community.
TOP
Rutidosis leptorrhynchoides
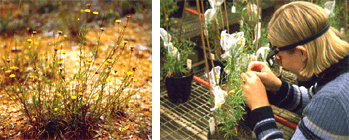
Rutidosis leptorrhynchoides (the button wrinklewort) has been a major study species for the last six years. R. leptorrhynchoides is a small perennial herb in the Asteraceae that is now only found in 26 populations ranging in size from 5 - >100 000 plants. The species is genetically interesting because it is self-incompatible (Young et al. 2001) and has two different chromosome races: 2n=22 for northern populations, while in the south of the range populations can also be 2n=44 autotetraploids (Murray & Young 2002). Some individuals with base numbers of n=13 also appear both as diploids and tetraploids.
Analysis of genetic diversity and mating patterns indicates that diploid populations smaller than 200 plants and isolated by more than 5 km show both reduced allozyme variation (Young et al. 1999) and increased levels of correlated paternity (Young & Brown 2000). High correlated paternity appears to be due to paternal bottlenecking as a result of erosion of S allele richness at the self-incompatibility locus (Young et al. 2001). These small populations also exhibit low seed set and high interplant variance in female fitness. This is reflected in reduced long-term viability as assessed using demographic matrix models based on six years of demographic monitoring data. The possible role of biparental inbreeding in further contributing to low population viability is currently being examined in a three generation crossing experiment - as recent work indicates that limited seed dispersal leads to the buildup of substantial spatial family structuring within populations (Wells & Young 2002).
Current research on R. leptorrhynchoides is focusing on the effects of outbreeding depression over a range of spatial scales. Of particular interest is how fitness reductions associated with outbreeding may trade off against the demographic limitations imposed by S allele mediated mate limitation in small genetically isolated populations. This is particularly important for this species with regard to re-establishment of new populations in the southern part of the species geographical range. Ecological factors determining individual plant-to-plant variation in female fitness such as floral display, spatial position, phenology and local mate density are also being examined (Young & Clemens).
TOP
Swainsona recta

Swainsona recta is a small purple pea found in fragmented grassland habitat where there are 17 known populations ranging in size from 10 to >400 reproductive plants. It is long-lived (>30 years) and, in contrast to R. leptorrhynchoides, is self-compatible. It is also an autotetraploid. Studies of allozyme variation and mating system (conducted using AUTOTET – Thrall & Young 2000) show strong reductions in both genetic variation and outcrossing as population size is reduced. However, in contrast to R. leptorrhynchoides, seed set does not decline in small populations, but germination and seedling fitness are reduced (Buza & Young 2000).
Comparison of these two grassland herbs provides a good example of how differences in breeding system mediate the ways in which genetic effects influence demography and viability, with R. leptorrhynchoides being limited by S allele erosion, while the dominant effect for S. recta is inbreeding depression. Similar research is underway on the herbs Rutidosis leiolepis (Young et al. 2002), Leucochrysum albicans (Costin et al. 2001) and the shrub Grevillea iaspicula (Hoebee & Young 2002).
Scientific Staff: Brown, Brubaker, Thrall, Young
TOP
Genetic and Ecological Viability of Plant Populations in Remnant Vegetation
This project has recently been established (September 2001) as a collaborative research effort between CSIRO Plant Industry, Conservation and Land Management (CALM) WA and the Native Vegetation Research and Development Program managed by Land and Water Australia. The objective is to extend previous genetic and ecological research on factors affecting the viability of rare plant populations to assessment of the role of genetic processes in determining the long-term viability of populations of common species occupying remnant vegetation. This is a critical step in developing integrated landscape management proposals aimed at conserving regional biodiversity as these more common species often provide much of the diversity and structural complexity of remnant vegetation that makes it useful as habitat for other species.
The underlying rationale is that persistent vegetation remnants must be made up of viable populations and that quantitative analysis of how landscape structure affects key biological processes for a modest number of target taxa, covering a range of life histories, will provide power to predict long-term persistence of broad classes of similar organisms under a range of remnant vegetation conditions.
Specific aims are to: 1) undertake quantitative analysis of ecological and genetic factors that affect the viability of plant populations in vegetation remnants; 2) determine how these are influenced by landscape attributes such as remnant size or connectivity; 3) assess how life history affects the impact of remnant characteristics on population viability and; 4) based on 1-3 develop alternative landscape design principles for major plant life history types that will maximise the probability of population persistence.

Three ecosystems are being studied: the temperate grasslands and grassy woodlands of southeastern NSW and the ACT, the kwongan shrublands and heathlands of Western Australia and the Brigalow woodlands of southern Queensland. Study species include herbs (e.g. Swainsona sericea) shrubs (e.g. Eremaea pauciflora; Calothamnus quadrifidus) and trees (e.g. Eucalyptus wondoo, E. aggregata and Acacia dealbata).
Similar research is also being conducted in collaboration with Prof. Brian Murray and Dr Gabi Schmidt-Adam at the University of Auckland in New Zealand on fragmented populations of the coastal broad-leaf tree pohutukawa (Metrosideros excelsa) (Schmidt-Adam et al. 1999; Young et al. 2001) and with Dr John Clemens at Massey University on Kakabeak (Song et al. 2002).
Scientific Staff: Broadhurst, Young
TOP
Modelling Effects of Self-incompatibility on Plant Population Viability
Theoretically, genetically controlled self-incompatibility systems may impose significant demographic constraints on small plant populations through loss of S allele richness and reduced mate availability. Such constraints represent one of the few direct links between genetic diversity and population viability that are of immediate importance for plant conservation. Significant loss of S alleles has recently been demonstrated in small plant populations (Young et al. 2000). In this research project, spatially explicit individual-based simulation approaches are being used to pursue two lines of investigation.
The first is a general analysis of the demographic and genetic consequences of different self-incompatibility systems for plants that vary in their reproductive capacity and lifespans. Results so far (Thrall et al. 2002) support the idea that, in the absence of inbreeding effects, populations of self-incompatible species will often be smaller and less viable than self-compatible species, particularly for shorter-lived organisms (high death rates) or where potential fecundity (ovule production) is low. While possessing some form of self-incompatibility reduces population size and persistence for a broad range of conditions, our results show that the actual number of S alleles is important for a more limited set of life-histories. In these situations increasing population viability through the addition of new S alleles may be the most effective approach to conservation. Comparison of model results to empirical data on disassortative mating provides a method for estimating S allele numbers in wild populations and assessing long-term effects of current mate limitation.
We are also undertaking a specific analysis of the long-term consequences of S allele limitation for the viability of populations of endangered species, using both empirical and modelling approaches. Here work is focusing on determining under what circumstances demographic vs genetic rescue are more appropriate management strategies for the herb Rutidosis leporrhynchoides and the bird pollinated shrub Grevillea iapsicula.
Scientific Staff: Thrall, Young
